
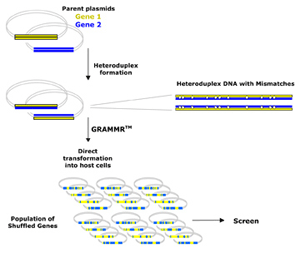
GRAMMR (Genetic ReAssortment by MisMatch Resolution) harnesses maximum genetic diversity and rapidly directs desired trait evolution for optimal product performance and gene expression.
Advantages:
- Iteration: successive rounds of selection evolve towards the desired trait rapidly with no sequence analysis and re-design needed
- Compatibility: Quickly re-assort libraries that have already been created into new combinatorial variants for screening
- Diversity: include diversity in regions that would not normally be targeted because they are outside the presumptive active site; let evolution decide what works!
Applications:
- Trait Evolution: enzymes can be evolved toward your desired trait (ex. thermal tolerance, higher specificity, yield) through iterative rounds of GRAMMR
- Empirical codon optimization: every combinatorial possibility of codon usage can be assessed through directed evolution (not limited by machine learning from imperfect data sets)
How it Works:
Click here to learn more about how GRAMMR works for directed evolution workflows.

